A cell is an information system#
A cell can be thought of as an information system with DNA, RNA, and proteins as the key players. Let’s look at some of the ways by which information is passed from molecule to molecule.
DNA → DNA#
Before a cell divides, it needs to prepare an entire new copy of genome. This is done by the process of replication. Differences between prokaryotes and eukaryotes (Write me).
DNA → RNA#
There are segments of DNA called genes, whose sequence information is used to synthesize RNA molecules. (As a side note, biology uses the word gene to mean so many different things. See [Gerstein et al., 2007] for the evolution of this term.) The biological process by which this happens is called transcription. As with replicatoin, there are difference in how this happens in prokaryotes vs. eukaryotes
Prokaryotes#
A key molecule that carries out transcription is called RNA polymerase. Here is a microscope image of a bacterial gene being transcribed. Notice how many RNA molecules are being synthesized from a single DNA molecule.
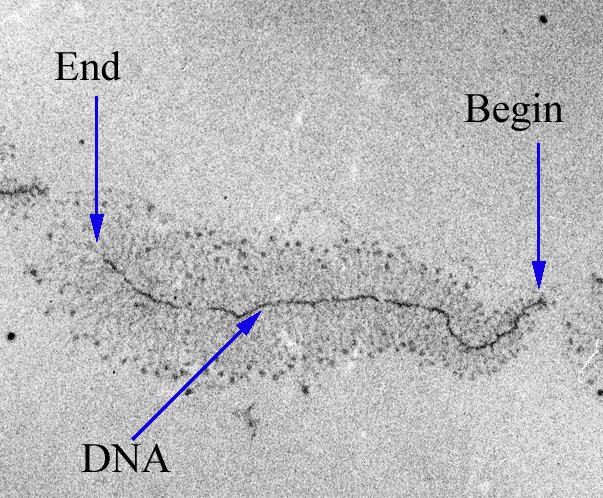
Fig. 8 Microscope image of transcription in action
Source: https://commons.wikimedia.org/wiki/File:Transcription_label_en_%28cropped%29.jpg#
Here’s a cartoon depicting transcription which shows what happens at the sequence level. The DNA sequence is simply copied into an RNA sequence.
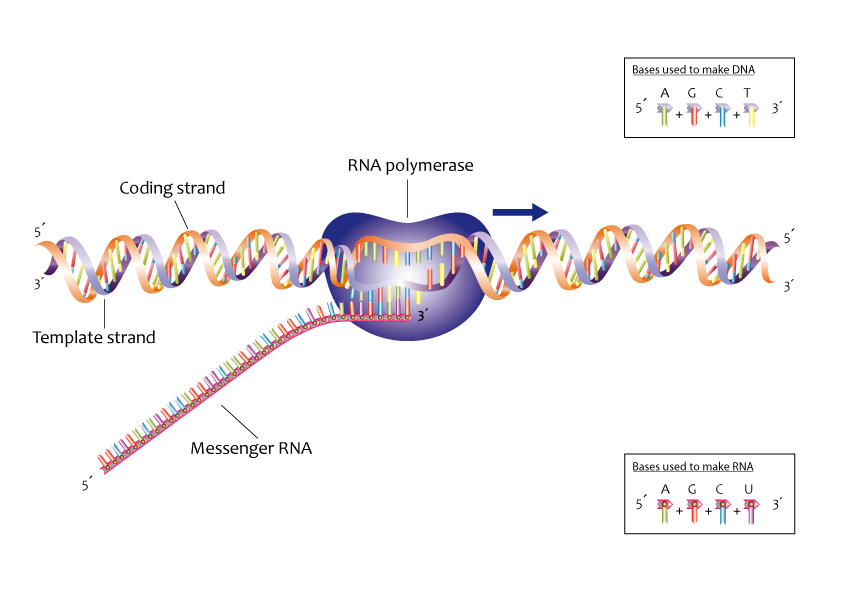
Fig. 9 Transcription zoomed in to sequence level
Source: https://commons.wikimedia.org/wiki/File:Transcription_label_en_%28cropped%29.jpg#
Eukaryotes#
In eukaryotes, DNA is first transcribed into premature RNA or precursor RNA or nascent RNA. Different to prokaryotes, three types of RNA polymerases are employed depending on the gene being transcribed. Also, since eukaryotic DNA is organized inside its nucleus, transcription happens inside the nucleus.
The nascent RNA then undergoes splicing in which some segments of the polynucleotide chain is removed and some other are retained. The ones removed are called introns and the ones retained are called exons. The resulting molecule is called a mature RNA.
To complicated matters further, the same nascent RNA molecule can give rise to a slightly or entirely different mature RNA depends on the choice of introns and exons. This is called alternative splicing. See Fig. 12 below.
RNA → Protein#
Transcription results in different kinds of RNAs being synthesized. Of these, non-coding RNAs go on to carry out whatever functions they have as RNAs. On the other hand coding RNAs (or messenger RNAs or mRNAs) merely hold sequence information which is used for the synthesis of proteins by a process called translation. A key player in translation is the molecule called ribosome.
Here’s a microscope image of bacterial translation in action (simultaneously with transcription). Can you guess what is what in the image?
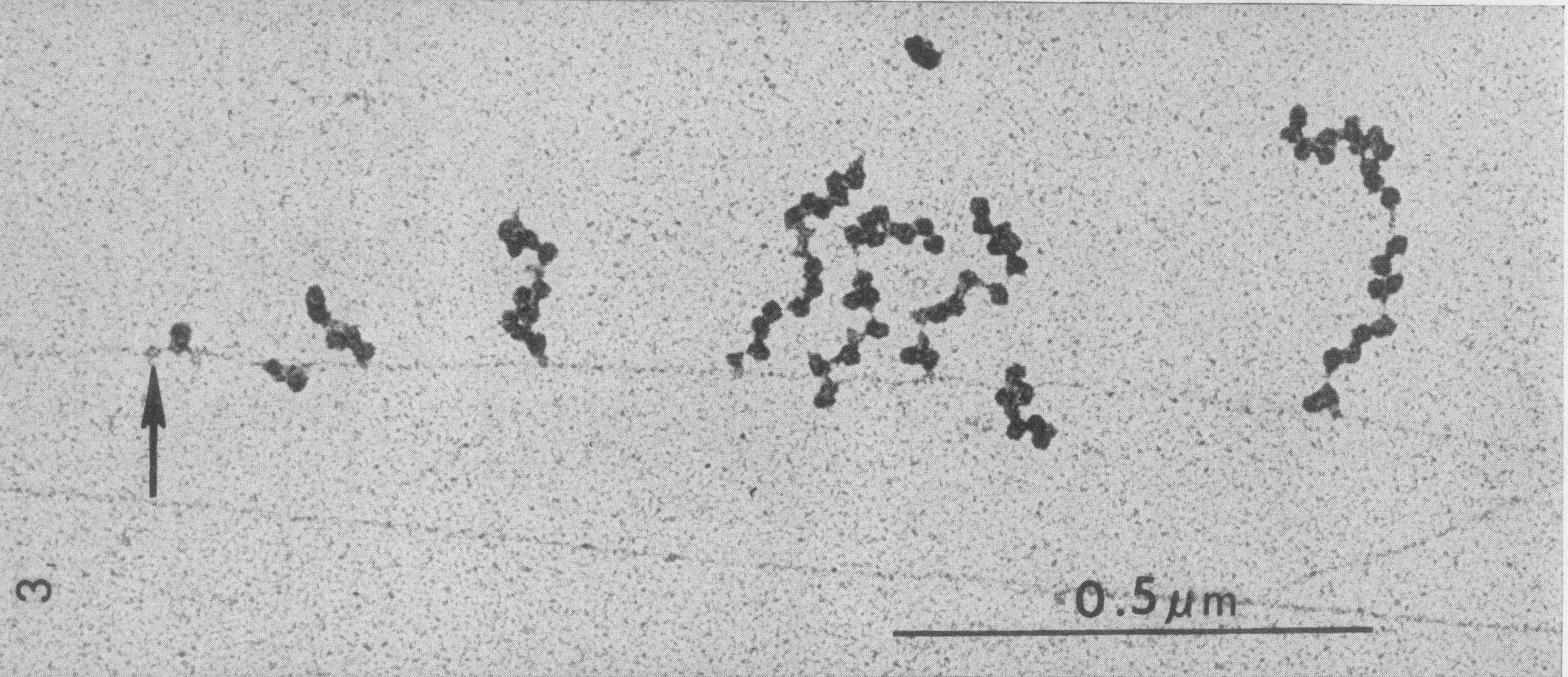
Fig. 10 Microscope image of translation and transcription
Source: Picture taken from [Miller et al., 1970] , O. L., Hamkalo, B. A., & Thomas, C. A. (1970). Visualization of Bacterial Genes in Action. Science, 169(3943), 392–395. doi:10.1126/science.169.3943.392#
Here’s a cartoon showing what happens at the sequence level during translation. Three nucleotides of mRNA, called a codon, get translated into an amino acid in the growing protein molecule.
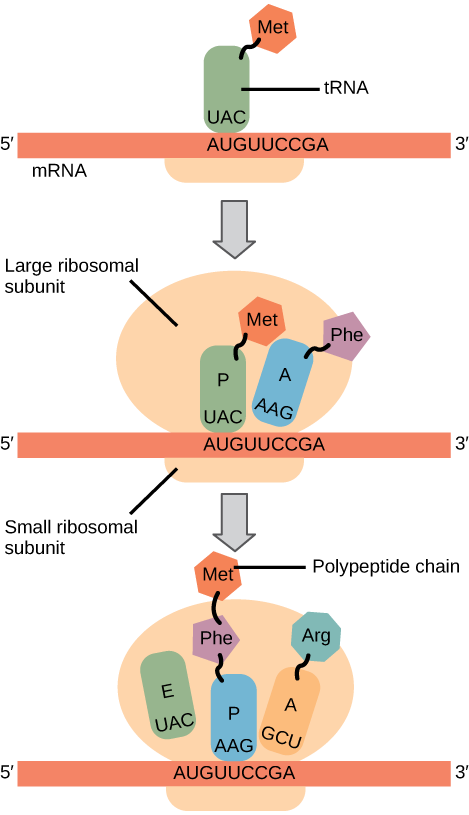
Fig. 11 Translation at the sequence level Source: {cite} Miller, O. L., Hamkalo, B. A., & Thomas, C. A. (1970). Visualization of Bacterial Genes in Action. Science, 169(3943), 392–395. doi:10.1126/science.169.3943.392#
In eukaryotes, alternative splicing means that a single gene can give rise to variant proteins which are referred to as protein isoforms.
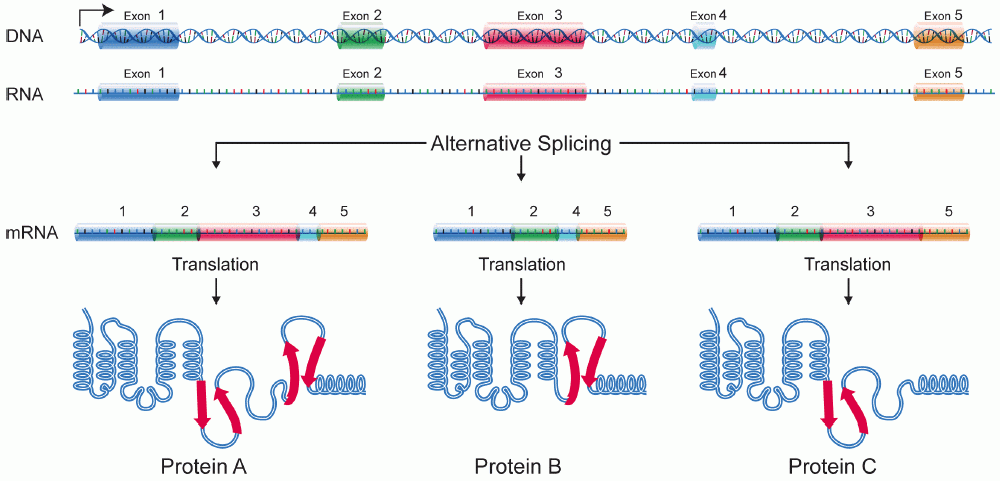
Fig. 12 Splicing and alternative splicing Source: https://en.wikipedia.org/wiki/Alternative_splicing#/media/File:DNA_alternative_splicing.gif#
The genetic code specifies which codon is translated to which amino acid#
So then, what is the correspondence between codons and amino acids? It turns out there is a deterministic rule about which codon codes for which amino acid. This is called the genetic code, although it’s more of a look-up table than a code. The figure below shows the ‘’standard’’ genetic code. This is almost universal, but some deviations are known.
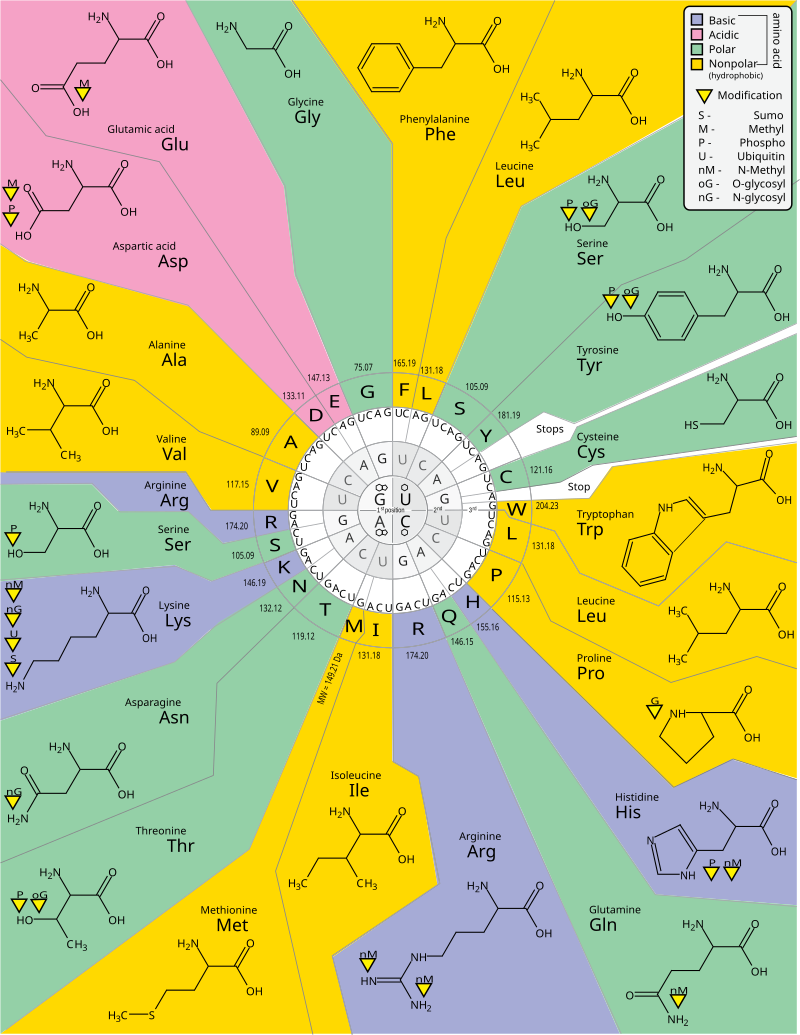
Fig. 13 Genetic code Source: https://en.m.wikipedia.org/wiki/File:GeneticCode21-version-2.svg#
How about these transfers: RNA → DNA or RNA → RNA or Protein → RNA or Protein → DNA?#
Back in 1950s when people were still trying to figure out what kinds of roles various biomolecules play in a cell, the consensus view was that information is transferred from nucleic acids to proteins, but not from protein to nucleic acid or protein to protein. This concept was coined the central dogma of molecular biology.
The dogma was not too dogmatic, as it was updated to allow for RNA → DNA transfers, which was previously not known to happen. This does happen a lot however, for example, when HIV-1 virus, which has RNA as its genetic material reverse transcribes it into DNA which then is integrated into the host genome. Another example is that of human telomere extension by an enzyme called telomerase reverse transcriptase. Experimental techniques for measuring RNA abundance or sequencing RNA also artifically carry out reverse transcription. Remember all the RT-PCR tests for SARS-Cov-2 you were forced to take during the pandemic? Anyway, the updated version of the still postulated that Protein → RNA, Protein → DNA, Protein → Protein transfer never occur.
Today we know a lot more about molecular genetics, and that RNA, DNA, proteins interact in many ways influencing the way in which their sequences are determined. So there is a need to revisit the central dogma ([Shapiro, 2009]), and while we are at it, perhaps give it a better name.
Further reading#
Replication, transcription, translations are all complex processes and highly regulated by many different factors. See [From DNA to RNA, n.d.] and [From RNA to protein, n.d.] for a more detailed treatment.
References#
From dna to rna. https://www.ncbi.nlm.nih.gov/books/NBK26887/.
From rna to protein. https://www.ncbi.nlm.nih.gov/books/NBK26829/.
Mark B Gerstein, Can Bruce, Joel S Rozowsky, Deyou Zheng, Jiang Du, Jan O Korbel, Olof Emanuelsson, Zhengdong D Zhang, Sherman Weissman, and Michael Snyder. What is a gene, post-ENCODE? history and updated definition. Genome Res., 17(6):669–681, June 2007.
O L Miller, Jr, B A Hamkalo, and C A Thomas, Jr. Visualization of bacterial genes in action. Science, 169(3943):392–395, July 1970.
James A Shapiro. Revisiting the central dogma in the 21st century. Ann. N. Y. Acad. Sci., 1178(1):6–28, October 2009.
