Hands-on: What genes are differentially expressed in colorectal cancer?#
Background#
Colorectal cancer (CRC) is one of the most commonly diagnosed cancers and one of the leading causes of cancer deaths (see e.g. this paper for more statistics). At the molecular level, many genes which otherwise play important roles in growth, damage repair, etc. act in abnormal ways in CRC (or any cancer for that matter). With RNA-seq, we can identify genes which exhibit differences in expression levels between normal and cancer tissues.
Objective#
The objective of this hands-on assignment is to identify differentially expressed genes in colorectal cancer using publicly available matched normal-cancer expression data.
Tasks#
Download gene expression data from the TCGA data portal. Filter by TCGA-COAD project. There are 41 normal samples and 476 tumor samples. Download at least 10 samples of each group. (If you want to retrieve more data, it might be faster to get programmatic access through their API or use a library like TCGAbiolinks handles it for you.)
The figure below shows count data for one of the samples.
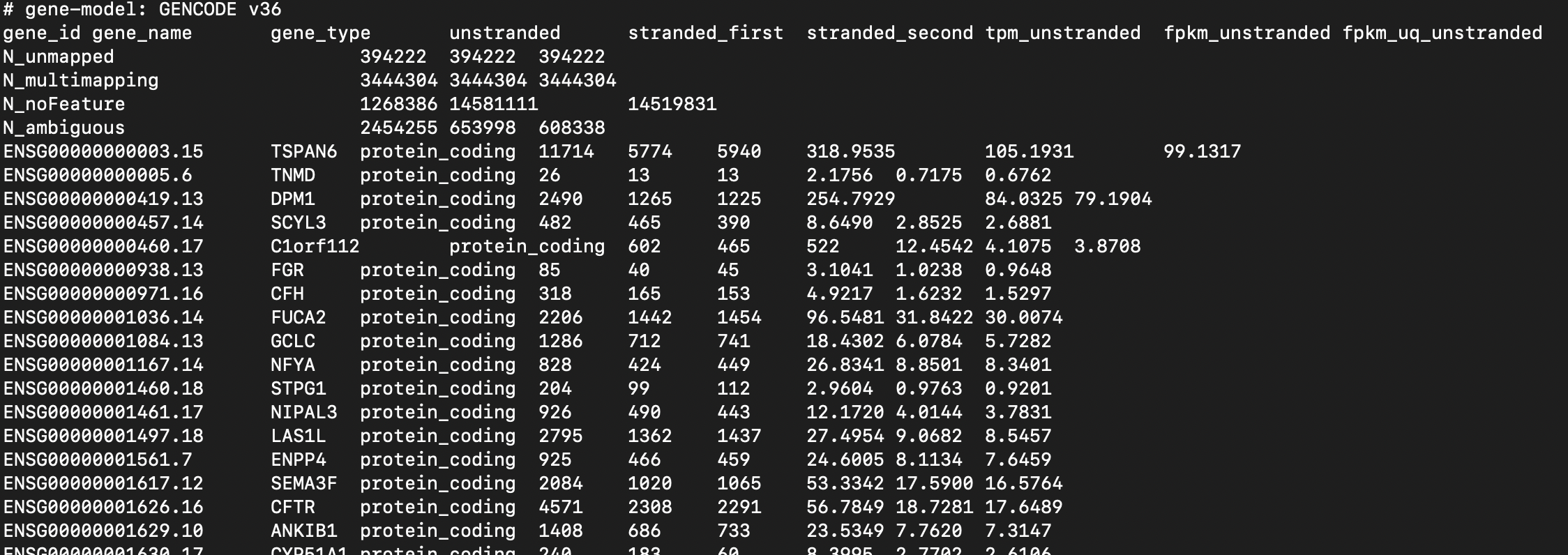
Fig. 49 A sample of gene expression data TCGA-COAD. A row corresponding to the measurements for a single gene. You can use the unstranded counts for further analysis.#
We discussed two kinds of approaches for modeling RNA-seq expression data. Below are two software tools represent. You can use either of the two.
limma-voom uses a linear model with transformed and variance-weighted expression values
DESeq2 uses a generalized linear model that assumes a negative binomial distribution for the expression data.
Prior to differential analysis, do some exploratory analysis. For example, check if the tumor and normal sample separate on a PCA plot based on log of depth-normalized counts.
Perform differential expression analysis using one of the tools above.
You will likely find > 1000 genes that are differentially expressed. Choose 10 genes based on p-value or the degree by which the expression has changed (reported as log fold change). Describe these genes.
References and useful links#
A practical guide to limma: (https://doi.org/10.12688/f1000research.9005.3)
A practical guide to DESeq2: https://bioconductor.org/packages/devel/bioc/vignettes/DESeq2/inst/doc/DESeq2.html
